Genomic Technologies in the Beef Industry
Genomics studies the structure, function, evolution and mapping of DNA and the genetic makeup of animals. It deals with the complete set of genes and genetic material found in a cell or organism.
Genomic technologies draw producer interest and research investment in the beef industry. Seedstock selection is one common application, but genomics has found widespread adoption in forage and feed grain breeding, diagnostic tests, vaccine development, source attribution for food safety recalls and other uses.
| Key Points |
|---|
| DNA is the genetic code that determines how an organism grows, what it looks like, and how it performs in a specific environment |
| DNA is a long chain, with each link of the chain containing a pair of four small molecules, known as base pairs or nucleotides. These molecules are abbreviated by the letters A, T, G, and C. This long chain is then coiled tightly into chromosomes. All cells in an organism contain a complete copy of that organism’s full genetic code. |
| Sometimes when DNA is copied, small copying mistakes occur. A “T” may be accidentally replaced with an A, G, or C. These are called single nucleotide polymorphisms (SNPs) and are regularly referred to as “markers” or “SNP markers.” |
| These SNPs largely dictate the physical differences we see among animals, including liveweight, stature, coat color, horns, marbling, or average daily gain, among others. |
| Even though an animal’s genetics are determined at conception, many environmental factors will modify how genes are expressed. |
| There are a number of ways that DNA testing can be used on an operation, including as an aid in bull and replacement selection, breeding choices, sorting animals into management groups, pedigree verification, eliminating or reducing undesirable genetic conditions, or marketing. |
Genetics 101
- Genetics: The study of individual genes and how they behave (heredity)
- Genomics: The study of how genes interact with each other and their environment and how the influence on phenotype
- Phenotype: the observable traits that result from an animal’s genotype interacting with the environment (e.g., coat colour, feed efficiency, horned/polled etc.)
DNA is the genetic code that determines how an organism grows, what it looks like, and how it performs in a specific environment. Found in all living things, DNA gets passed from one generation to the next, allowing these organisms to maintain or improve their ability to survive and thrive.
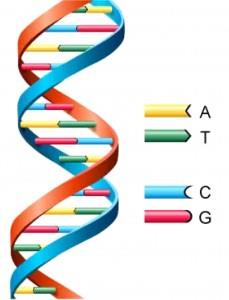
DNA is a long chain, with each link of the chain containing a pair of four small molecules, known as base pairs or nucleotides. These molecules are abbreviated by the letters A, T, G, and C. This long chain is then coiled tightly into chromosomes. All cells in an organism contain a complete copy of that organism’s full genetic code.
Each cell has specialized machinery that reads the DNA code three letters at a time. These three-letter codes instruct the cellular machinery to start “reading” at a specific point. From that point, sets of three base pairs code for specific amino acids, then finally a three-letter code instructs the cellular machinery to stop reading. This segment – from start to stop – is referred to as a gene.
The amino acids produced by the coding instructions of genes then link together to create proteins.. Some genes code for specific proteins in the body, such as hormones, enzymes, plant proteins and muscle proteins. Other genes influence when specific genes are turned on or off, or how actively genes are expressed.
Sometimes when DNA is copied, small copying mistakes occur. A “T” may be accidentally replaced with an A, G, or C. These are called single nucleotide polymorphisms (SNPs) and are regularly referred to as “markers” or “SNP markers.” So far, beef geneticists have discovered around 30 million different SNPs. These SNPs largely dictate the physical differences we see among animals, such as live weight, frame size, coat color, horns, marbling, or average daily gain, among others. Identifying all 30 million of these SNPs in a single animal (i.e. whole genome sequencing) is expensive, therefore labs make smaller “panels” of these SNPs, which makes genotyping more affordable to use in animal breeding. One can categorize these panels into five different categories based on SNP density, with panels of higher density costing more:
- Small panels: analyze just a few SNPs to around 2,000 SNPs. Generally used for parentage, genetic abnormality testing, coat colour, horns, etc.,
- Low Density (LD) panels: contain between 5,000 to 30,000 SNPs. This type of panel is used for genomic prediction/selection purposes when there are a lot of higher density genotypes available to refer to.
- Medium Density (50K) panels: have between 50,000 and 250,000 SNPs. This density is fairly common in beef genomic selection programs. Usually, a population will begin by gathering a reference set of 50,000 SNP genotypes on influential animals in the breed, which are then used to predict what genotypes mean for performance traits.
- High Density (HD) panels: analyze 500,000 to 1 million SNPs. These panels are typically used in research and are not widely used in commercial settings.
- Whole Genome Sequencing (WGS): occurs when an animal’s entire genome is sequenced (i.e., all 3 billion base pairs are read). This type of test is presently used primarily for research and is too expensive to be employed on farm.
Even though an animal’s genetics are determined at conception, many environmental factors can modify how genes are expressed. For example, an animal may have a very high genetic potential for weight gain, but if there is a drought or feed is limited, the animal will be unable to fulfill that genetic potential. Some traits follow a simple inheritance pattern, meaning the presence of one gene will dictate whether or not an animal has the trait (e.g. horned or polled). Other traits require a combination of a few genes to see an effect, and even more traits have a complex inheritance pattern in which multiple genes as well as the environment all interact to produce a phenotype.
Click below to download our genomics fact sheet:

The Use of Genomic Tests in Cattle
There are a number of ways that DNA testing can be used on an operation, including as an aid in bull and replacement selection, breeding choices, sorting animals into management groups, pedigree verification, eliminating or reducing undesirable genetic conditions, or marketing. Outside of the farm, DNA testing can be used to provide insight into evaluation, help us understand common and complex diseases, determine ancestral history, and used to analyze evidence in criminal investigations. Implementing DNA tests on an operation can be very valuable, but it is important to have a defined goal in mind before investing in testing. The value of DNA testing to an operation is dependant on breed, number of animals tested, whether cattle are purebred or commercial, breeding goals, marketing scheme, as well as what records are being kept on farm.
For example, if you sell your calves at weaning and are looking to DNA parentage test your calves to determine which sires are most economically valuable (or sire the most calves with high weaning weights), but are not recording individual calf weaning weights the test would be of limited value.

DNA samples for most genetic tests can be obtained through hair, skin tissue, blood, or semen. Sampling kits can be obtained from laboratories or third-party providers (i.e., breed associations) and will provide details on collecting the sample and what information needs to accompany the sample depending on the specific test ordered.
Genomically-Enhanced EPDs
Expected progeny differences (EPDs) are a prediction of the average differences in genetic merit that are expected to be passed on to the offspring. A full description of EPDs and how they can be used to make selection decisions on your herd can be found here.
When looking at EPDs, it is important to note the accuracy. In young animals, EPDs are relatively inaccurate as they have not produced any progeny. As more progeny are produced and more information on offspring performance is available, the EPDs of the sire and dam become more accurate. Bull EPDs are usually more accurate than cow EPDs because the average bull has many more offspring than the average cow. Genomic tests can be used to increase accuracy of prediction without having to sire more calves. Genomically-enhanced EPDs (GE-EPDs) can be produced for most breeds in Canada. This increases the accuracy of the EPD by the same amount as having added performance information from seven to twenty-three offspring, depending on the specific trait. This allows producers to better be able to better predict the genetic merit of progeny from young animals (heifers or yearling bulls) before they have produced any calves.
Adding genetic information to create an EPD does not guarantee that the EPD itself will be more desirable, just that it will be more accurate. For example, a bull’s calving ease EPD may improve, worsen, or stay the same after a GE-EPD evaluation. The major difference is that whatever that actual EPD number is, a producer can have more confidence that the bull’s calving ease will be reflected by the EPD due to the higher accuracy.
For someone looking to purchase bulls or replacement heifers, GE-EPDs should be used in the exact same way EPDs are used. The accuracy value for the specific trait will help you determine how much variation to expect.
Selection Indexes
Selection indexes are a suite of EPDs that are directly related to a breeding objective weighted by their economic importance for that objective. Basically, indexes represent the breeding value of an animal as a parent based on how the selected EPDs will influence profit of their progeny. The index is computed as below:
The types of indexes are:
- Terminal Indexes
- Predict profit when progeny are fed and sold
- Includes production traits such as gain, carcass value, DMI and feed efficiency
- Maternal Indexes
- Predict profit when replacement females are retained, and steers and cull heifers are sold
- Includes traits such as weaning weight, milk, calving ease, mature cow size and longevity
- All purposed
- Terminal calves and replacement calves are retained (e.g., backgrounding)
- Includes both terminal and maternal traits such as weaning weight and carcass value
The fewer traits in an index allows for faster progress in the breeding objective. While there is concern with the ‘one size fits all’ assumption, indexes have been proven to be highly accurate across variable environments and management practices. There is also a lot of value in customizable indexes which are also available for commercial usage.
Indexes are most commonly expressed as the expected difference in profits between a parents’ progeny. There a few indexes that are standardized to 100, 100 being the average. Anything below 100 will be a below average performing bull for that particular index while anything above 100 is above average. Both of these methods are used to compare the expected progeny performance difference of bulls.
For example, if Bull A and Bull B were both evaluated using the same index:
Bull A = $300
Bull B = $185
This means on average, progeny from Bull A will be $115 more profitable than the progeny from Bull B for that specific breeding objective.
A number of Canadian breed associations and other organizations have selection indexes available for use by seedstock and commercial producers to help make breeding decisions.
DNA Parentage Testing
DNA parentage testing has been used in the purebred industry for many years and is now being adopted by commercial cow-calf producers as prices for the tests drop. Generally, a parentage panel contains 120 SNPs used to determine which bull sired which calf. The present cost of parentage using a 120 SNP parentage panel is $12-$20.
Producers might adopt DNA parentage testing on their operations for several reasons. They may need to verify the sire for purebred registry requirements or for parentage discovery if cows are bred in multi-sire pastures. Producers may want to identify a ‘problem’ bull on their operation, such as a bull that sires high birth weight calves, low weaning weight calves, or fewer calves than expected. If the right records are kept on farm, parentage testing can help identify superior sires (i.e., bulls siring high weaning weight calves or calves with a high ADG), identify sires that are producing calves with desired traits, or sires that are not breeding many cows.
Parentage verification SNP tests take advantage of the fact that half a calf’s DNA is inherited from the sire and half from the dam. The tests look at specific SNPs from both the calf and all possible sires. A process of elimination then determines the sire – if the calf carries a SNP not carried by a particular bull, that bull can be excluded as the sire. Therefore, it’s important to submit samples from all possible sires. Sometimes, especially in cases where potential sires are closely related, DNA from only the sire and calf is not enough to narrow down which bull sired the calf and a sample from the dam may be required.
Trait Assessment
An animal that has inherited the same version of a particular gene from each parent is called homozygous. If an animal receives a different form of the gene from each parent, it’s called heterozygous. Heterozygous animals can pass on either form of the gene to their offspring,
SNP tests can be used to determine if animals are “carriers” for a specific trait. Recessive traits require that the offspring inherit a copy of the gene from each parent before the recessive trait is expressed. Otherwise, the dominant copy from one parent (i.e., polled) will mask the recessive copy from the other parent (i.e.,., horned). SNP tests can help to unmask carriers for some traits.
One example of a simply inherited dominant/recessive trait is coat colour. For example, in black cattle, red hair is the recessive form of the gene, so even if an animal is black, it may still carry a red gene, and could pass the red gene to its calves. If two carriers are bred to each other, there is a one-in-four chance their offspring will express the recessive trait even though neither of the heterozygous parents do. Following is an example showing what happens when mating a cow and bull that are both heterozygous black.
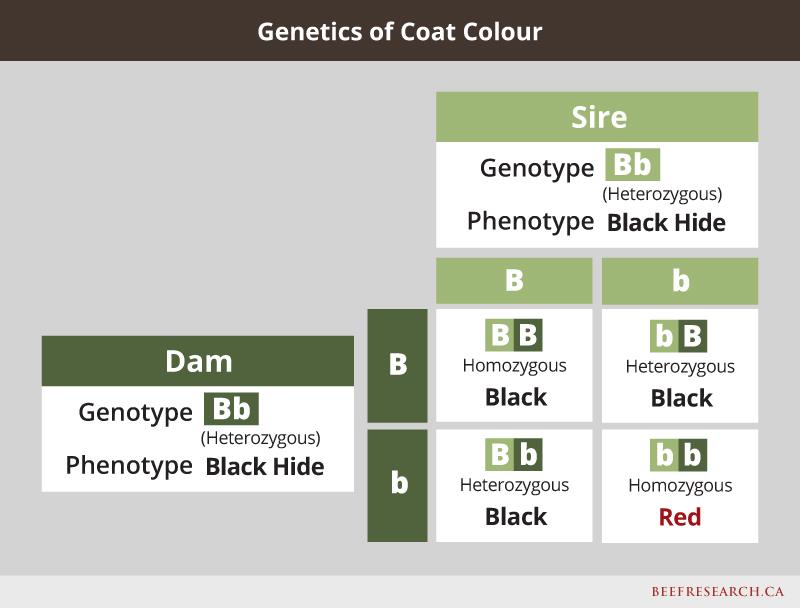
Past research funded by the Canadian Beef Cattle Check-off validated that a mutation in the calpastatin gene was associated with differences in beef tenderness and is controlled by this simple inheritance pattern.
Genetic Abnormality Testing
Abnormalities in the skeleton, body formation, or the way the body functions occasionally occur in beef cattle. They can be caused by the environment (i.e., crooked calf disease caused by lupine consumption), genetics, or both. If environment plays a factor, adjustments can be made to prevent the disease in the future. If the disease is genetic, testing may be required to correct or manage the problem.
Some examples of genetic abnormalities are: Chondrodysplasia (dwarfism), Hypotrichosis (hairlessness), Arthrogryposis Multiplex (Curly Calf Syndrome) and Syndactyly (mulefoot).
Genetic abnormalities are often breed specific, only occur in a few breeds, or only occur in specific bloodlines within a breed. Some breed associations are working to reduce the frequency of genetic defects within their breeds by using genetic testing to identify carriers. Most genetic abnormalities follow a simple dominant/recessive inheritance pattern, so you can avoid genetic abnormalities by avoiding mating carriers to each other.
Genetic abnormalities can result in abortion, or death shortly after birth. If you suspect that you are seeing a genetic abnormality in your herd, talk to your veterinarian to determine what’s causing the defect. If it’s genetic, test your bulls for carrier status. Avoid buying bulls that are carriers for the defect, or crossbreed to a different breed that doesn’t have the abnormality.
Genetic Tests to Sort Animals into Management Groups

Genetic tests can be used to manage groups of cattle. Tests can give producers a better idea of how animals will perform in specific situations. These tests enable producers to sort animals into particular management groups. This is commonly referred to as marker assisted management (MAM).
For example, the leptin gene codes for the hormone leptin that regulates appetite and fat deposition. For this particular gene, cattle will have a base pair code of CC, TC or TT. The TT calves will typically deposit backfat faster and require fewer days on feed than TC or CC calves. By leptin testing, feedlot operators can sort calves into more uniform groups to feed and sell.
Recently a genetic test has been developed that allows producers to know the breed composition (or heterozygosity) of an individual animal, which can help producers with future breeding decisions if they wish to maximize heterosis.
Cautions Regarding the use of Genetic Tests
Genomics works very well for parentage testing, controlling inbreeding, and testing for many genetic abnormalities. However, caution must be exercised when using genomics for selection or management. When traits are controlled by a single gene, selection is relatively simple., But single trait selection for any one trait is not advised, as selecting for only one trait often means selecting for or against other traits unintentionally. Most production traits (i.e., growth or fertility) are controlled by hundreds of different genes that all interact with each other, which makes those traits challenging selection targets. For example, calves from a homozygous polled dam will always be polled because they are guaranteed to inherit at least one copy of the polled gene. But, if a dam had a short calving interval every year, it is not guaranteed that her progeny will display that same short calving interval because there are several different genes that influence calving interval, and it is unlikely that all of her calves will inherit all of the genes that produce the most desirable result.
While genetic testing works very reliably in cases where the SNP is known to occur within an actual gene (i.e. functional or causative mutation), in many cases the SNP may only be located somewhere near the gene. In those instances, the accuracy of GE-EPDs will depend on the relationship between the animals tested and the population from which the prediction equations used to generate the GE-EPDs were developed. GE-EPD prediction equations using SNPs that were discovered in one bloodline may not work as reliably in another, and GE-EPD prediction equations developed using SNPs that were discovered in one breed are unlikely to produce reliable results in a different breed. When using genetic predictions, it is important to use ones developed for populations that are or are closely related to the population in which the prediction was created for (e.g., use Charolais indexes for your Charolais herd). Since the beef cattle herd primarily consists of crossbred animals, the inability to provide SNP markers that reliably predict crossbred animal performance has been a major barrier to the adoption of genomic selection in the commercial cattle population.
Genomics in Animal Health
Diagnostic Tests
Numerous diagnostic tests use genomics. Advancements in genomics have improved the ability to detect and understand many animal diseases.
Because researchers can now map the genome of a specific virus or bacteria, detecting them in a host animal becomes easier. For example, genomics helped develop improved tests for trichomoniasis (trich) and bovine genital campylobacteriosis (vibrio). The trich test can detect pathogenic trichomonads and ignore harmless trichomonads that may also be present, while the vibrio test improved upon traditional culture methods that often resulted in false negatives. Using genomic technologies, more rapid, accurate and cost-effective diagnostic tests are now available.
Researchers also use genomic technologies to improve current diagnostic tests. Genomics in diagnostics is faster and just as reliable as culture-based methods. Research funded through the BCRC is developing a single, rapid, cost-effective test that can detect a range of respiratory and enteric diseases.
Disease Outbreak and Food Safety Investigations
In the case of disease outbreaks, genomics and genetic testing of the causative agent can play a major role. Scientists are able to take a sample from an infected animal and map the genome of the causative bacteria. By examining the genetic sequence of the pathogen, it may be possible to determine the origin of the infection, if multiple strains are involved (suggesting multiple routes of infection or transmission) and can help to inform control strategies or policy.
Genomics has facilitated major advancements in the area of traceability. In the event of a food safety recall, genetic tests can be used to link bacterial DNA to the processing facility and identify where along the production chain the contamination occurred.
Compared to bacterial culture, genomic testing increases both the accuracy and the speed with which food safety outbreaks can be traced.
Vaccine Development
Genomics can also be used to develop more effective vaccines, discovering new infectious agents, and providing a better understanding of known vaccines and pathogens.
Vaccines encourage a cow’s immune system to develop antibodies against specific disease organisms. An important part of vaccine development is finding a unique protein on the bacteria or virus (antigens) that the animal’s immune system recognizes and then stimulates the immune response.
Genomic technology can help to identify the best proteins to stimulate immunity and makes it much easier to multiply and produce these proteins in large quantities. This increases the effectiveness of the vaccine by making it more specific, with fewer side effects. It also decreases the cost of producing the vaccine.
Genetic Improvements in Feed Efficiency
Feed costs represent 50-70% of costs on a beef operation. Reducing feed inputs can have huge advantages to your bottom line. This can be done through improving feed efficiency. A 5% improvement in feed efficiency could have an economic effect four times greater than a 5% improvement in average daily gain (ADG). There are several tools available to genetically select for feed efficiency.
Measuring Feed Efficiency
Residual Feed Intake (RFI) Testing
If you ask an average cattle producer how they measure feed efficiency in their herd, most would say by looking at the feed to gain ratio (F:G) or the feed conversion ratio (FCR). However, this is typically measured using herd, group or pen data resulting in a group/herd/pen average. Residual feed intake is one of many tools that can be used to estimate individual feed efficiency.
Residual feed intake is defined as the difference between an animal’s actual feed consumed, or eaten, and the animal’s calculated feed requirements based on its body weight and ADG during a standardized test period. Essentially, RFI describes the variation in feed intake that remains after the requirements for maintenance and growth have been met. Efficient animals eat less than expected and have a negative or low RFI, while inefficient animals eat more than expected and have a positive or high RFI.
More recently, RFI values have been adjusted for body fatness (RFIfat), thus attempting to render RFI independent of carcass fatness in slaughter cattle and later maturity or fattening in replacement heifers and bulls. Similarly, another measure referred to as residual gain (RG) is adjusted for body size and dry matter intake (DMI). It represents animals with superior gain at equal levels of body weight and DMI. A trait that combines both RFI and RG, referred to as residual intake and gain (RIG), represents metabolically efficient, fast-growing animals that consume less feed. These latter two measures of feed efficiency should also be adjusted for body composition using final off-test ultrasound backfat thickness, marbling score and/or ribeye area. These measures of feed efficiency are heritable and either moderately (RFI vs. RG) or highly (RG vs. RIG) correlated with each other. However, there are other measures of efficiency being explored. Likely, there probably isn’t one method that will be appropriate for every operation.
Selecting for Feed Efficiency
Seedstock producers and have effectively used expected progeny differences (EPDs) to improve the genetic merit of their cattle. However, it is important to avoid single trait selection. Balancing trait selection for the inputs and not just the outputs is critical to optimize profitability on beef operations. In any population, there is variation in feed efficiency, and there will be individuals that can achieve high rates of gain with low feed intakes just as there will be individuals achieving low rates of gain with high feed intakes and others in between. The challenge comes in measuring cattle for this trait, interpreting the numbers and using the data for genetic progress.
Figure 4 shows the difference in average DMI in control cattle and those selected for RFI efficiency. The below graph shows 4 years of data in which the Efficient group had lower DMI than the Control group to an extent that approaches significance P = 0.095. The Efficient herd also had a lower average EBV for RFI of 0.026±0.015 Kg DMI/d in comparison to 0.044±0.017 Kg DMI/d in the control herd. The assumption can be made that extending this study will show RFI selected cattle have a lower DMI while maintaining efficient growth to a significant extent. As seen in previous studies. Selecting for high feed efficient cattle can result in reduced feed costs and potential environmental benefits in the form of reduced GHG emissions.
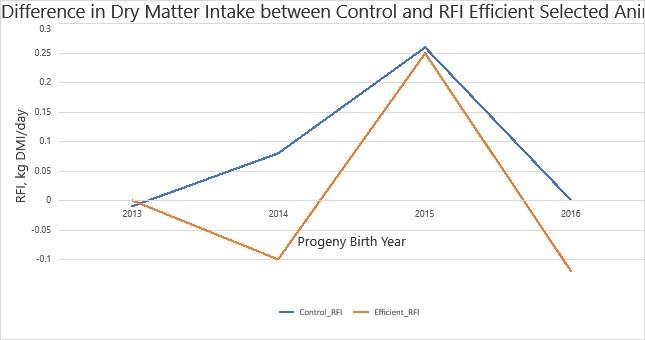
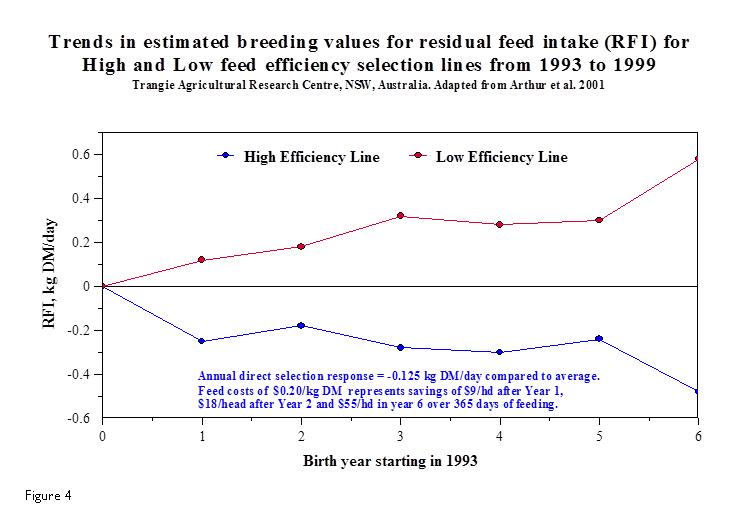
Figure 5 compares the progress that has been made in dry matter intake and the resulting feed costs throughout the 1990’s at an Australian Research Centre in regards to actual selection for feed efficient cattle by using RFI. The negative RFI (high efficiency cattle) have reduced their dry matter intake per day by close to 0.5 kg resulting in significant feed savings to the beef operation. However, given that multi-trait breeding goals will be pursued by the industry, the researchers assumed an annual rate of response in RFI of -0.08 kg DM/day (0.8%/year) in a 25-year simulation compared to not selecting for RFI. Much progress can be made in improving feed efficiency well into the future as each generation improves after targeted selection for the trait.
Currently, the easiest way to select for feed efficiency measured by RFI (if you are not testing your own cattle) is to purchase RFI bulls to use on your cow herd. Typically, RFI is measured in young cattle (7-10 months of age) in feedlot pens fitted with feeding stations designed to automatically monitor individual animal feed intake over a 70-day test (e.g., Figure 5. GrowSafe Systems Ltd., Airdrie, Alberta, Canada) following a three-week adjustment to the test diet. Cattle are weighed before feeding on two consecutive days at the start and end of the test period and at approximately 14-28 day intervals. They are also measured for ultrasound backfat thickness (mm), rib eye area (cm2) and marbling score at the start (optional) and end of the test period. Even though we have a great tool to measure feed efficiency in the RFI trait, it should be balanced with other traits of economic value. For a cow-calf producer, fertility is 10 times more important than carcass traits and 5 times more important than growth traits. Single trait selection is never a wise decision, nor a practical goal to pursue.
Impact of Selecting for Low RFI
Several researchers around the world, including Dr. John Basarab, a Research Associate in the Faculty of Agricultural, Life and Environmental Science at the University of Alberta, have been studying this trait for more than 15 years. They have come to the following conclusions about what selection for feed efficient cattle means to the beef industry.
Research has shown that selection for low RFI (efficient cattle) will:
- Have no effect on growth, carcass yield, or quality grade
- Reduce feed intake at equal weight and ADG
- Improve feed to gain ratio by 10-15%
- Reduce net energy of maintenance and reduce methane and manure production (reducing the carbon footprint of cattle)
- Have a slight if any effect on age at puberty (~ 3 days)
- Have no effect on calving pattern in first calf heifers
- Have no negative effect on pregnancy, calving or weaning rate
- Have little effect on bull fertility
- Have a positive effect on body fatness or weight in mature cows particularly during stressful periods
- Will reduce feed costs
- $0.07-0.10/hd/d feeders
- $0.11-0.12/hd/d in cows

By looking at the cows below, you likely have a preference for one over the other when their feed efficiency isn’t known. When you find out they both weaned calves of similar weights and they both were pregnant, you are probably inclined to find the deep bodied, more feminine looking cow on the right as more appealing. When we look at RFI scores and see this same cow ate 2.83 kg more per day (6.2 lbs) than the average cow in the herd and 5.4 kg (11.9 lbs.) than the other cow (Figure 7), you may reconsider knowing the impact on feed costs for the entire herd.. While these examples are extreme, they illustrate how conformation alone may not explain differences in productivity or performance. Granted, these are large cows, but the concept of feed efficiency is the same for other breeds and sizes of cows. The argument has been made by producers that their 1100 lb mature cows are all efficient because they are smaller. Remember, RFI is independent of body size and weight, average daily gain and backfat thickness and variation and extremes exist in all populations.

Relationship Between Feed Efficiency and Other Important Production Aspects
Many factors affect actual feed intake (DMI) of cattle such as body size, growth, body composition, gender, age, season, ambient temperature, physiological status, previous nutrition and diet. Most of these factors are either equal between animals during a standardized feed intake test (e.g., gender, season, ambient temperature and physiological status) or are adjusted for, such as body size, body composition, and growth so that we can make direct comparisons between animals. However, considerable within- and between-animal variation does exist in DMI and measures of feed efficiency.
Diet Type and Breed Type
RFI measured on a grower diet and then again on a finisher diet is positively correlated, meaning the animals tend to rank similarly on both diets when tested. This is often true of RFI measured post-weaning in heifers and RFI measured again later in life as mature cows. However, some animals do re-rank, meaning some will have a positive RFI (+RFI) or be inefficient in one test during a specific production or growth phase and on a specific diet, but when tested again under different parameters are efficient or have a negative RFI (-RFI). This level of re-ranking for RFI, DMI and ADG can occur if the diet changes from a grower to finisher diet or even when the diet stays the same from feeding period to feeding period. Re-ranking was due to: 1) body weight and feed intake measurement error, 2) animal variation in response to compensatory gain, 3) animal variation in efficiency with animal maturity and, 4) animal variation in diet digestibility due to differences in feeding behavior, rate of passage and rumen microbial population.
Preliminary data from the Lacombe Research Centre, Canada, demonstrated moderate to strong repeatability of RFI over different stages of the animal’s life. Replacement heifers identified as -RFI and +RFI when they were 8-12 months of age and fed a 90:10% barley silage and barley grain diet (as fed; -0.373 vs. 0.365 kg DM/day) were also -RFI and +RFI when measured again as 4–7-year-old cows and fed a 70:30% grass hay and barley straw cube diet (as fed; -0.375 vs. 0.459 kg DM/day).
There may be a need for different test criteria (e.g., forage vs. grain-based diets) when selecting terminal and maternal bulls. Breed types destined for maternal purposes should be tested on forage diets whereas those breed types designed for terminal sire purposes should be tested on grain-based diets.
Body Size and Carcass Traits
RFI is not related to pre- and post-weaning growth, body size, slaughter weight and carcass traits in beef cattle and the phenotypic and genetic correlations are near zero. Experiments have shown cattle with negative RFI values tend to have less backfat but no difference in carcass weight, dressing percentage, or marbling grade compared to cattle of positive RFI values. However, in an Australian study, the muscle of efficient animals was found to be slightly leaner and also have slightly more calpastatin than inefficient steers, which may negatively affect meat tenderness. Tenderness is being monitored in efficient animals to see if these results are repeatable in other cattle populations. RFI is moderately to highly correlated with feed intake and feed conversion ratio (FCR). This implies that selection for negative RFI will decrease feed intake at equal levels of body weight, growth and body fatness, and will improve feed-to-gain ratio in feeder cattle and growing replacement heifers.
Methane Emissions
The fact that efficient cattle have decreased feed intake at equal levels of body weight, growth and body fatness also implies that selection for low RFI will decrease methane (CH4) emissions (g/animal/day) because methane emissions are proportional to feed intake. Generally, the higher the DMI, the higher the methane emissions are. These estimated reductions in methane emissions of 9-12% also coincide with a 15-17% reduction in manure production. Improvements to feed efficiency will influence the carbon footprint of cattle making beef more sustainable.
After 25 years of selection for –RFI in an Australian simulation model, the efficient 120-cow beef herd had lower GHG emissions by 101 t CO2e per year or 0.844 t CO2e per cow per year compared with the average 120-cow herd. In addition, the carbon footprint of the efficient herd was 14% lower than that of the cow herd not selected for RFI (19.82 vs. 23.06 kg CO2e/kg carcass beef) and the total farm area decreased by 13%. These estimates of GHG reduction are conservative as improved DM digestibility in –RFI cattle and improved accuracy and rate of genetic change resulting from genomic enhanced breeding values were not considered.
Feeding Behaviours and Temperament
Feeding behaviours that are collected as part of the RFI test suggest that inefficient (+RFI) cattle use about 5% more energy in feed related activities and possibly spend less time ruminating. They may also be more reactive to stressors since they visit the feed bunks (9-22%) more often each day and feed more often at night. Inefficient cattle may also be less docile and more nervouscompared to efficient cattle.
Body Condition and Adaptability
Observations of beef cows in extensive Canadian winter conditions has shown that –RFI (efficient) cows maintain themselves in better body condition score with no differences in productivity compared with their +RFI herd mates. Dams that produced -RFI progeny consistently had 2-3 mm more back fat thickness, on average, over the 12th and 13th ribs and lost less weight during early lactation (pre-calving to pre-breeding) than mothers that produced +RFI progeny. In addition, –RFI (efficient) heifers calving for the first time had lower calf deaths within 30 days of birth than +RFI (inefficient) heifers. Lower calf death loss suggests that the improved early life survival of calves from -RFI mothers may be due to their improved feed efficiency resulting from more available nutrients and a better uterine environment compared with +RFI mothers. Recent research from the Lacombe Research Centre confirm these findings in that –RFI cows gained more body fat and body weight than +RFI cows when both groups swath grazed forages for the first time during Canadian winters where night time temperatures dropped below -20° C and animals grazed through the snow from November to March. Previous to swathgrazing, both –RFI and +RFI young cows had been wintered together in smaller holding areas and fed barley silage to meet their nutritional requirements. Even though efficient cattle have documented lower feeding event duration and frequency and lower feed intake when bunk fed in an RFI test, this does not mean that –RFI animals cannot compete or acquire forages during extensive grazing. Instead, it may imply that efficient animals are more adaptable and less susceptible to stress than +RFI or inefficient animals.
Fertility and Productivity
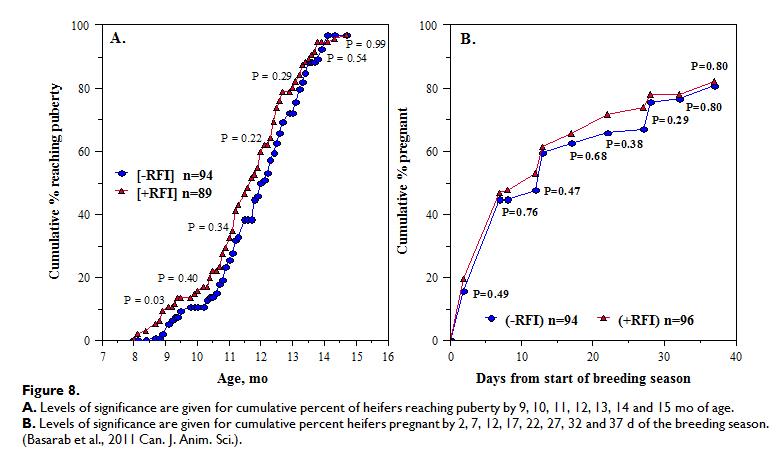
The relationships between RFI and fertility and productivity in heifers and cows have recently been reviewed, showing that -RFI and +RFI cows and heifers were similar in culling, pregnancy, calving and weaning rate, calving pattern and kilogram of calf weaned per mating opportunity (in both Canadian and Australian data). However, -RFI (efficient) cows calved 5-6 d later in the year than +RFI (inefficient) cows. When RFI was adjusted for body fatness (final off-test backfat thickness; RFIfat) no differences were observed in percentage of -RFIfat and +RFIfat heifers reaching puberty by 10, 11, 12, 13, 14 or 15 months of age nor in the percentage of calves born by day 28 of the calving season. Calving difficulty, age at first calving, calf birth weight, calf pre-weaning ADG, calf actual and 200-d weaning weight and heifer productivity, expressed as kg calf weaned per heifer exposed to breeding, were also similar between -RFI and +RFI heifers. Figure 8 below shows how –RFI (efficient) heifers appear to reach puberty later and get pregnant later in the breeding season compared to +RFI (inefficient) heifers.
Bull Fertility
Fertility of young bulls, as measured by scrotal circumference, breeding soundness evaluation, calves born per sire and semen characteristics, have mostly been unrelated to RFI, though several weakly negative associations have been observed with sperm morphology and motility in negative RFI bulls. These observations provide another reason to adjust RFI for ultrasound backfat thickness and feeding behaviors in an effort to prevent the selection for later maturing bulls. A study at the University of Alberta Kinsella herd showed there was no difference in the number of calves sired by –RFI bulls compared to +RFI bulls. We have to be careful to not link sperm morphology alone with infertility since other aspects of a breeding soundness evaluation are important to ensure any bull, regardless of RFI status, is acceptable to be put into a breeding situation.
Data continues to be collected from collaborative research projects focused on feed efficiency and residual feed intake both in Canada and internationally. Careful interpretation of the data when applying or adopting the technology to your beef operation is required. While there is large variation in the range of RFI values in animals, more than 35%, and because the trait is moderately heritable, significant genetic progress can be achieved in breeding programs, resulting in cost saving benefits. However, with time and continued improvement in technologies, the ability to detect efficient animals will improve and increase our ability to select superior animals.
Plant Breeding

Plant breeders have been using genomics for years to help select valuable traits. Understanding a plant’s genetic sequence allows plant breeders to combine genomic tools with traditional breeding methods. That helps shorten the time it takes to develop new varieties and makes it easier to select for complex traits.
The use of genomic technology allows plant breeders to develop varieties with characteristics that are more robust, disease resistant, higher yielding, cold tolerant, winter hardy and so forth. Genomics allows plant breeders to identify superior lines more quickly and accurately, and make more strategic selection and breeding decisions, resulting in more rapid genetic progress.
Current research funded by the Canadian Beef Cattle Check-off is using genomics as a tool to develop improved tame and native forages.
Future Potential
Our understanding of genomics continues to evolve. Ongoing research on the genetic improvement of beef cattle utilizing genomic testing includes whether we can select for improved disease resistance or maternal production traits to inform replacement heifer selection. These areas continue to evolve, but other areas related to the beef industry continue to advance at an even more rapid pace. Genetic traceability is now possible, and some value chains are marketing full farm to plate traceability through DNA sampling. New genomic methods of plant breeding are being used and developed, and any future chute side disease diagnostic platforms will likely incorporate genomics. As our understanding of genomics and genetics grows, the potential to use these technologies to benefit multiple aspects of beef production continues to expand.
- References:
-
Berry D.R., Farcia J.F., Garrick D.J. 2016. Development and implementation of genomic predictions in beef cattle. Animal Frontiers. 6:1 32-38
Van Eenennaam A.L. 2015. How might DNA-based information generate value in the beef cattle sector? UC Davis extension.
Perez-de-Castro A.M., Vilanova S., Canizares J., Pascual L., Blanca J.M., Diez M.J., Prohens J., Pic. 2012. Application of genomic tools in plant breeding. Curr Genomics. 13:3 179-195
Raszek M.M., Guan L.L., Plastow G.S. 2016. Use of genomic tools to improve cattle health in the context of infectious diseases. Front. Genet. 07
Bergen R. 2012. Getting a grip on genomics. Canadian Cattlemen’s Magazine. July 2012 edition
Bergen R. 2012. More on genomics. Canadian Cattlemen’s Magazine. September 2012 edition
Feedback
Feedback and questions on the content of this page are welcome. Please e-mail us.
This content was last reviewed April 2022.
